Identifying and targeting the correct cells to treat diseases like Rheumatoid Arthritis (RA) is now cheaper and more accessible thanks to a new 3D printed droplet microfluidic control instrument.
Developed by researchers from New York Genome Center (NYGC) and New York University (NYU), the instrument is open source, fits on a bedside and can be obtained and assembled for around $600, which is 20 to 200 times cheaper than other comparable instruments.
“Most commercial microfluidic instruments are very costly; as a result, not every lab has access to exciting technology for single-cell analysis,” said Dr. William Stephenson, who led the study and the development of the instrument. “We designed the instrument to perform droplet microfluidics and in particular Drop-seq, a massively parallel technology for single cell RNA-sequencing.”
Manufacturing the control instrument
While 3D printing has been used to manufacture the microfluidic vessels themselves and in 3D bioprinting, NYGC only used the technology here as part of the experimental set-up.
The instrument is comprised of electronic and pneumatic components affixed to a 3D printed frame. The entire system is operated through software control using a graphical user interface on a touchscreen.
To manufacture the instrument, a multi-tiered 3D printed frame was FFF 3D printed, measuring 21 cm by 20 cm, with a height of 9 cm. The frame includes a mount for the raspberry pi chip and a tablet computer. For the fluid handling, commercially available micro air-pumps, regulators, and micro solenoid valves were used.
A shaft was 3D printed and added to a permanent magnet stepper motor to stir barcoded microparticles used in the cell analysis, which could then be visualized on the screen.
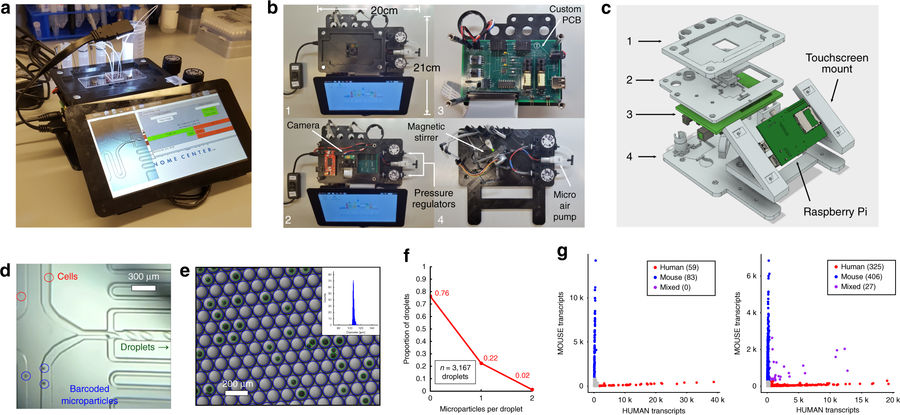
Analysing cells with the microfluidic control instrument
The researchers used the instrument to profile joint synovial tissue from RA patients, an autoimmune disease that affects 1% of the US population. They collected samples from five RA patients totaling 20,387 cells and looked at the individual gene expression patterns for each cell.
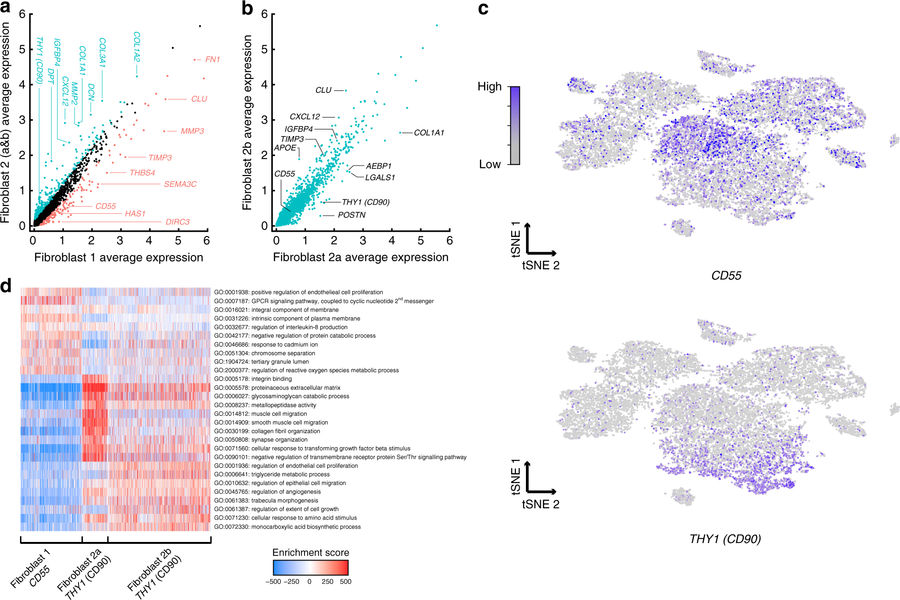
By analyzing the complete dataset and searching for clusters of similar cells, the researchers identified distinct groups of fibroblasts (connective tissue cells) with strikingly different gene expression patterns, an important step towards creating a comprehensive “cell atlas” for synovial tissue from RA patients. Researcher and lead author Rahul Satija said:
“This dataset gave us the opportunity to identify individual subpopulations of cells that could drive the progression of RA, even if they have not been previously characterized.”
“From this work, we have classified unrecognized fibroblast subtypes that may prove to be important drug targets for our RA patients,” added fellow lead author Laura Donlin.
Other potential applications
Other potential applications of this system include recent work profiling immune repertoires from hundreds of thousands of single cells, hydrogel microsphere fabrication for 3D cell culture, chemical microfluidic gradient generation, and microparticle size sorting.
The full study, Single-cell RNA-seq of rheumatoid arthritis synovial tissue using low-cost microfluidic instrumentation by William Stephenson, Laura T. Donlin, Andrew Butler, Cristina Rozo, Bernadette Bracken, Ali Rashidfarrokhi, Susan M. Goodman, Lionel B. Ivashkiv, Vivian P. Bykerk, Dana E. Orange, Robert B. Darnell, Harold P. Swerdlow and Rahul Satija is available to read online via nature magazine.
Want to design the trophy for this year’s awards? Protolabs is sponsoring the 2018 3D Printing Industry Awards design competition. Submit your design now to win a 3D printer.”
For more stories on 3D printing and microfluidics research, subscribe to our free 3D Printing Industry newsletter, follow us on Twitter, and like us on Facebook.
Featured image shows the microfluidic control instrument attached to a tablet. Photo via NYGC.


